2. model Package¶
This package provides two kinds of models:
- Generative pointprocess models (see Section ni.model.pointproccess)
- GLM based inhomogeneous pointprocess (ip) models
The pointprocess models can instantiate a given firing rate function with a spike train. The ip models use “components” that are combined to fit a spike train using a GLM.
2.1. What is a “component”?¶
For the ni.model package, a component is a set of time series (or a way to generate these) in a Generalized Linear Model that is used to fit a set of spike trains. When a model is fitted to a set of spike trains, the components are used to compute the designmatrix. Each component provides 1 or more timeseries that represent the influence of a specific event or aspect that is expected to affect the firing rate at that specific point in time to a certain degree.
Eg. a component can be as simple as a constant value over time, or it can be 0 for most of the time, except at those time points where a specific event occurs (eg. a spike of a different neuron). The design matrix shown here is mostly 0 and contains three components (history, rate and a constant):
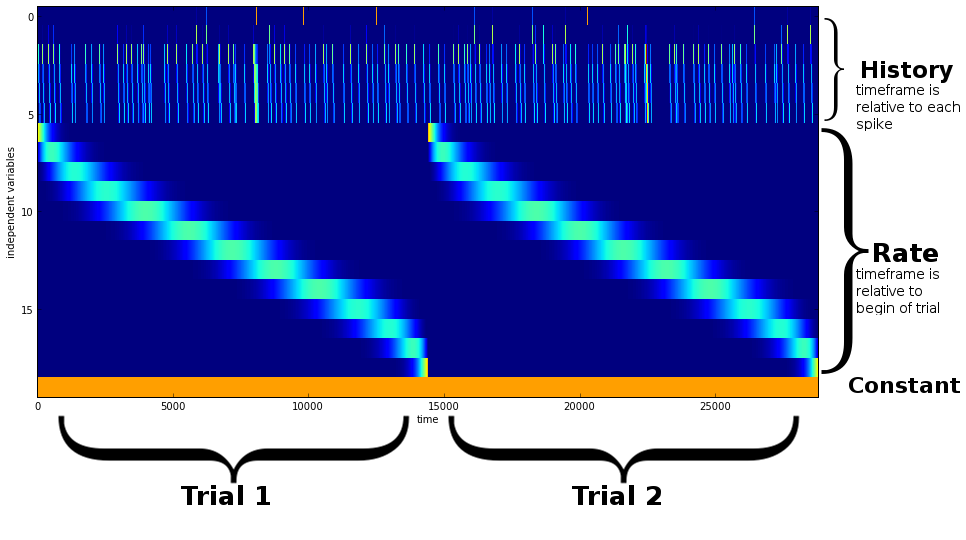
To model a more or less precise time course of the effect of an event, the component needs to create mutiple time series, each representing a time window relative to the event (usually following the event, if a causal link is assumed). It is convenient to use splines to model these windows, because they will produce a smooth time series when summed. These time windows can be arranged linearly (each spanning the same amount of time) or logarithmically (such that there is a higher resolution close to the event than further away). The length and number of the time windows can be adjusted, since their explanatory value depends on the modeled process.
2.2. The Rate component¶
Given that each spike train is alligned in the same timeframe regarding a certain stimulus (or sequence of stimuli), a rate component models rate fluctuations that occur in every trial. This is done by creating a number of timeseries, each representing a portion of the trial time: eg. early in the trial, in the middle of the trial, at the end of the trial.
A ni.model.designmatrix.RateComponent will span the whole trial with a given number of knots while the class ni.model.designmatrix.AdaptiveRateComponent will use the firing rate to space out the knots to cover (on average) an equal amount of spikes.
2.3. The history components¶
Since the spike data containers contain multiple spike trains, a component can access the spiketimes of each of the spike trains contained. The history components use these spike times to model the effect of a certain spike train on the dependent spike train.
- autohistory: the effect of the spike train on itself. Eg. refractory period or periodic spiking
- crosshistory: models the effect of another spike train on the one modeled
- 2nd order autohistory_2d: for bursting neurons, the autohistory alone will not capture the behaviour of the neuron, as it has to either predict periodic activity for the whole trial or no auto effect at all. The 2nd order history takes into account two spikes, such that the end of a burst can be predicted.
Here you can see the slow time frame of a rate fluctuation (fixed to the time frame of each trial) and the fast time frame of a history component (each relative to a spike):
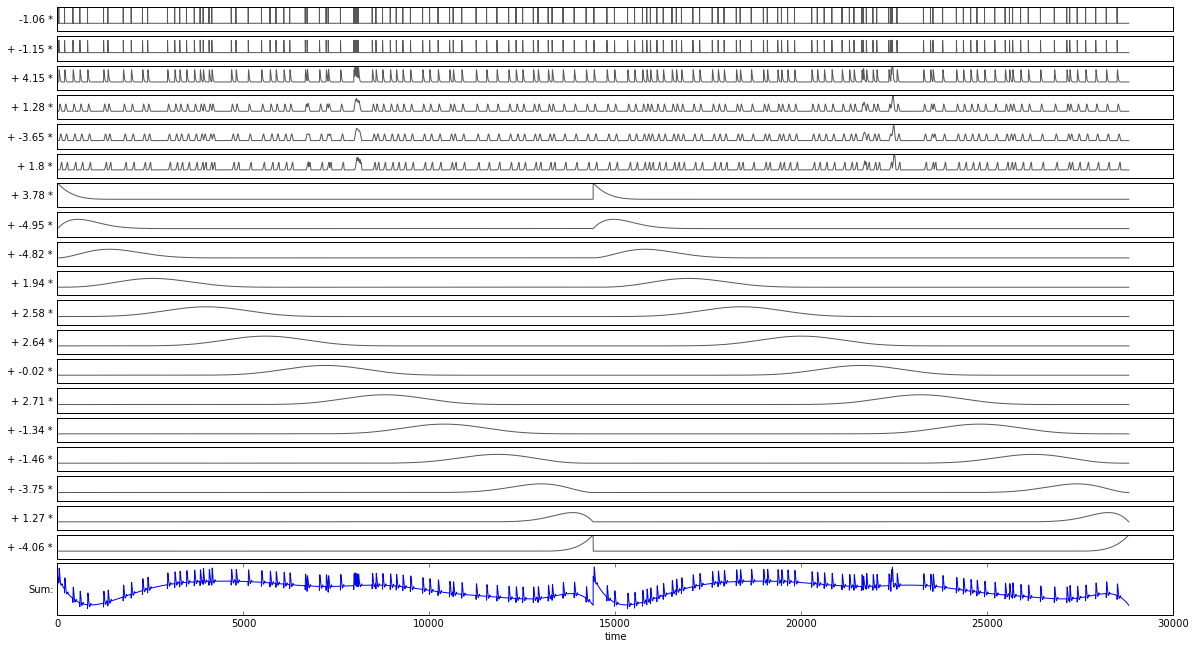
2.4. Configuring a Model¶
The model class has a default behaviour that assumes you want to have one rate component, one autohistory componenet and a crosshistory component for each channel. This behaviour can be changed by providing a different configuration dictionary to the model.
Eg. to create a model without any of the components:
model = ni.model.ip.Model({‘name’:’Model’,’autohistory’:False, ‘crosshistory’:False, 'rate':False})
Or, if additionally the 2nd order autohistory should be enabled:
model = ni.model.ip.Model({‘name’:’Model’,’autohistory_2d’:True })
Important for models is also which spike train channel should be modeled by the others:
model_0 = ni.mode.ip.Model({'cell':0})
model_1 = ni.mode.ip.Model({'cell':1})
model_2 = ni.mode.ip.Model({'cell':2})
model_3 = ni.mode.ip.Model({'cell':3})
Instead of only true or false, crosshistory can also be a list of channels that should be used:
model = ni.mode.ip.Model({'cell':1,'crosshistory':[2]}) # cell 1 modeled using activity of cell 2
Also there are a number of configuration options that are passed on to a component to change its behaviour:
- Rate Component:
knots_rate: how many knots should be created
- adaptive_rate: Whether the RateComponent or AdaptiveRateComponent should be used. The AdaptiveRateComponent can additionally be tweaked with:
- adaptive_rate_exponent (default = 2)
- adaptive_rate_smooth_width (default = 20)
- History Components (autohistory and crosshistory):
- history_length: total lenght of the time spanned by the history component
- history_knots: number of knots to span
These options are set for the whole model at once:
model = ni.mode.ip.Model({'knots_rate':10,'history_length':200}) # sets history_length for autohistory and crosshistory, knots_rate for rate component
2.5. Fitting a Model¶
An ip.Model can be fitted on a ni.data.data.Data object and will produce a ni.model.ip.FittedModel. This will contain the fitted coefficients and the design templates, such that the different components can be inspected individually:
model = ni.model.ip.Model({'cell':2,'crosshistory':[1,3]})
fm = model.fit(data.trial(range(data.nr_trials/2)))
fm.plot_prototypes() # will plot each fitted component
plot(fm.firing_rate_model().sum(1)) # will plot the firing rate components (rate + constant)
plot(fm.statistics['pvalues']) # plots the pvalues for each coefficient
2.6. Creating custom components¶
To be precise, what happens when a model is fitted is the following:
- The model creates a ni.model.designmatrix.DesignMatrixTemplate and adds components (which are subclasses of ni.model.designmatrix.Component) according to its configuration to it:
- when the Component classes are created they are provided with most of the information they need (eg. trial length)
- The DesignMatrixTemplate is combined with the data which creates the actual designmatrix by:
- For each component the function getSplines is called, providing the component with spikes of other neurons
- The component returns a 2 dimensional numpy array that has the dimensions of length of time x number of time series
- if the array does not have the length of the complete time series, it will be repeated until it fits. So a kernel that has the length of exactly one trial will be repeated for each trial (the ni models by default make all trials the same length).
The design matrix is then passed to the GLM fitting backend
So, if you want to add a component, this can be either implement a function getSplines that returns a 2d numpy array that has the correct dimensions (time bins x N), or you can use or inherit from ni.model.designmatrix.Component and set the self.kernel attribute which will then be returned:
my_kernel = ni.model.create_splines.create_splines_linspace(time_length, 6, 0) # creates a 6 knotted set of splines covering time_length
c = Component(header='My own component',kernel=my_kernel)
model = ni.model.ip.Model(custom_components = [c])
Applications can be eg. a rate component that is only applied to trials, while a second component is applied to the others. If two kinds of trials are alternated this could be done like this:
from ni.model.designmatrix import Component
my_kernel = ni.model.create_splines.create_splines_linspace(time_length, 6, 0) # creates a 6 knotted set of splines covering time_length
is_trial_type_1 = repeat([0,1]*(number_of_trials/2),trial_length)
is_trial_type_2 = 1 - is_trial_type_1
c1 = Component(header='Trial Type 1 Rate',kernel=np.concatenate([my_kernel]*number_of_trials) * is_trial_type_1[:,np.newaxis])
c2 = Component(header='Trial Type 2 Rate',kernel=np.concatenate([my_kernel]*number_of_trials) * is_trial_type_2[:,np.newaxis])
model = ni.model.ip.Model({'custom_components': [c1,c2]},trial_length)
Or alternatively you can overwrite a kernel of the rate component:
kernel = np.concatenate([
np.concatenate([my_kernel]*number_of_trials) * is_trial_type_1[:,np.newaxis]),
np.concatenate([my_kernel]*number_of_trials) * is_trial_type_2[:,np.newaxis])
])
model = ni.model.RateModel({’rate’:True, ’custom_kernels’: [{’Name’:’rate’,’Kernel’: kernel}]})
2.7. Classes in the ni.model package¶
- class ni.model.BareModel(configuration={})[source]¶
Bases: ni.model.ip.Model
This is a shorthand class for an Inhomogenous Pointprocess model that contains no Components.
This is completely equivalent to using:
ni.model.ip.Model({‘name’:’Bare Model’,’autohistory’:False, ‘crosshistory’:False, ‘rate’:False})
- class ni.model.RateModel(knots_rate=10)[source]¶
Bases: ni.model.ip.Model
This is a shorthand class for an Inhomogenous Pointprocess model that contains only a RateComponent and nothing else.
This is completely equivalent to using:
ni.model.ip.Model({‘name’:’Rate Model’,’autohistory’:False, ‘crosshistory’:False, ‘knots_rate’:knots_rate})
- class ni.model.RateAndHistoryModel(knots_rate=10, history_length=100, history_knots=4)[source]¶
Bases: ni.model.ip.Model
This is a shorthand class for an Inhomogenous Pointprocess model that contains only a RateComponent, a Autohistory Component and nothing else.
This is completely equivalent to using:
ni.model.ip.Model({‘name’:’Rate Model with Autohistory Component’,’autohistory’:True, ‘crosshistory’:False, ‘knots_rate’:knots_rate, ‘history_length’:history_length, ‘knot_number’:history_knots})
2.7.1. ip Module¶
- class ni.model.ip.Configuration(c=False)[source]¶
Bases: ni.tools.pickler.Picklable
The following values are the defaults used:
name = “Inhomogeneous Point Process”
cell = 0
Which cell to model in the data (index)Boolean Flags for Default Model components (True: included, Fals: not included):
autohistory = True crosshistory = True rate = True constant = True autohistory_2d = False
backend = “glm”
The backend used. Valid options: “glm” and “elasticnet”backend_config = False
knots_rate = 10
Number of knots in the rate componentadaptive_rate = False
Whether to use linear spaced knots or an adaptive knot spacing, which depends on the variability of the firing rate. Uses these two options:
adaptive_rate_exponent = 2 adaptive_rate_smooth_width = 20
history_length = 100
Length of the history kernelknot_number = 3
Number of knots in the history kernelcustom_kernels = []
List of kernels that overwrite default component behaviour. To overwrite the rate component use:
[{ 'Name':'rate', 'Kernel': np.ones(100) }]
custom_components = []
List of components that get added to the default ones.self.be_memory_efficient = True
Whether or not to save the data and designmatrix created by the fitting modelUnimportant options no one will want to change:
delete_last_spline = True design = 'new' mask = np.array([True])
Look at the [source] for a full list of defaults.
- class ni.model.ip.FittedModel(model)[source]¶
Bases: ni.tools.pickler.Picklable
When initialized via Model.fit() it contains a copy of the configuration, a link to the model it was fitted from and fitting parameters:
FittedModel. fit
modelFit OutputFittedModel. design
The DesignMatrix used. Use design.matrix for the actual matrix or design.get(‘...’) to extract only the rows that correspond to a keyword.- compare(data)[source]¶
Using the model this will predict a firing probability function according to a design matrix.
Returns:
Deviance_all: dv, LogLikelihood_all: ll, Deviance: dv/nr_trials, LogLikelihood: ll/nr_trials, llf: Likelihood function over time ll: np.sum(ll)/nr_trials
- generate(bins=-1)[source]¶
Generates new spike trains from the extracted staistics
This function only uses rate model and autohistory. For crosshistory dependence, use ip_generator.
bins
How many bins should be generated (should be multiples of trial_length)
- plot_firing_rate_model()[source]¶
returns a time series which contains the rate and constant component
- plot_prototypes()[source]¶
plots each of the components as a prototype (sum of fitted b-splines) and returns a dictionary of figures
- class ni.model.ip.Model(configuration=None, nr_bins=0)[source]¶
Bases: ni.tools.pickler.Picklable
- compare(data, p, nr_trials=1)[source]¶
will compare a timeseries of probabilities p to a binary timeseries or Data instance data.
Returns:
Deviance_all: dv, LogLikelihood_all: ll, Deviance: dv/nr_trials, LogLikelihood: ll/nr_trials, llf: Likelihood function over time ll: np.sum(ll)/nr_trials
- dm(in_spikes, design=None)[source]¶
Creates a design matrix from data and self.design
in_spikes ni.data.data.Data instance
design (optional) a different designmatrix.DesignMatrixTemplate
- fit(data=None, beta=None, x=None, dm=None, nr_trials=None)[source]¶
Fits the model
in_spikes ni.data.data.Data instanceexample:
from scipy.ndimage import gaussian_filter import ni model = ni.model.ip.Model(ni.model.ip.Configuration({'crosshistory':False})) data = ni.data.monkey.Data() data = data.condition(0).trial(range(int(data.nr_trials/2))) dm = model.dm(data) x = model.x(data) from sklearn import linear_model betas = [] fm = model.fit(data) betas.append(fm.beta) print "fitted." for clf in [linear_model.LinearRegression(), linear_model.RidgeCV(alphas=[0.1, 1.0, 10.0])]: clf.fit(dm,x) betas.append(clf.coef_) figure() plot(clf.coef_.transpose(),'.') title('coefficients') prediction = np.dot(dm,clf.coef_.transpose()) figure() plot(prediction) title('prediction') ll = x * log(prediction) + (len(x)-x)*log(1-prediction) figure() plot(ll) title('ll') print np.sum(ll)
- generateDesignMatrix(data, trial_length)[source]¶
generates a design matrix template. Uses meta data from data to determine number of trials and trial length.
2.7.2. designmatrix Module¶
- class ni.model.designmatrix.AdaptiveRateComponent(header='rate', rate=False, exponent=2, knots=10, length=1000, kernel=False)[source]¶
Bases: ni.model.designmatrix.Component
Rate Design Matrix Component
header: name of the kernel component rate: a rate function that determines exponent: the rate function will be taken to thins power to have a higher selctivity for high firing rates knots: Number of knots length: length of the component. Will be multiplied
kernel: use this kernel instead of a newly created one
- class ni.model.designmatrix.Component(header='Undefined', kernel=0)[source]¶
Bases: ni.tools.pickler.Picklable
Design Matrix Component
header: name of the kernel component kernel: kernel that will be tiled to fill the design matrix
- class ni.model.designmatrix.DesignMatrix(length, width=1)[source]¶
Bases: ni.tools.pickler.Picklable
Use DesignMatrixTemplate to create a design matrix.
This class computes an actual matrix, where DesignMatrixTemplate can be saved before the matrix is instanciated.
- class ni.model.designmatrix.DesignMatrixTemplate(length, trial_length=0)[source]¶
Bases: ni.tools.pickler.Picklable
Most important class for Design Matrices
Uses components that are then combined into an actual design matrix:
>>> DesignMatrixTemplate(data.nr_trials * data.time_bins) >>> kernel = cs.create_splines_logspace(self.configuration.history_length, self.configuration.knot_number, 0) >>> design_template.add(designmatrix.HistoryComponent('autohistory', kernel=kernel)) >>> design_template.add(designmatrix.HistoryComponent('crosshistory'+str(2), channel=2, kernel = kernel)) >>> design_template.add(designmatrix.RateComponent('rate',self.configuration.knots_rate,trial_length)) >>> design_template.add(designmatrix.Component('constant',np.ones((1,1)))) >>> design_template.combine(data)
- class ni.model.designmatrix.HistoryComponent(header='autohistory', channel=0, history_length=100, knot_number=4, order_flag=1, kernel=False, delete_last_spline=True)[source]¶
Bases: ni.model.designmatrix.Component
History Design Matrix Component
Will be convolved with spikes before fitting
header: name of the kernel component channel: which channel the kernel should be convolved with (default 0) history_length: length of the kernel knot_number: number of knots (will be logspaced) order_flag: default 0 (no higher order interactions)
kernel: use this kernel instead of a newly created one
Atm only order 1 interactions
- class ni.model.designmatrix.HistoryDesignMatrix(spikes, history_length=100, knot_number=5, order_flag=1, kernel=False)[source]¶
Internal helper class
- class ni.model.designmatrix.RateComponent(header='rate', knots=10, length=1000, kernel=False)[source]¶
Bases: ni.model.designmatrix.Component
Rate Design Matrix Component
header: name of the kernel component knots: Number of knots length: length of the component. Will be multiplied
kernel: use this kernel instead of a newly created one
- class ni.model.designmatrix.SecondOrderHistoryComponent(header='autohistory', channel_1=0, channel_2=0, history_length=100, knot_number=4, order_flag=1, kernel_1=False, kernel_2=False, delete_last_spline=True)[source]¶
Bases: ni.model.designmatrix.Component
History Design Matrix Component with Second Order Kernels
Will be convolved with spikes before fitting
header: name of the kernel component channel: which channel the kernel should be convolved with (default 0) history_length: length of the kernel knot_number: number of knots (will be logspaced) order_flag: default 0 (no higher order interactions)
kernel: use this kernel instead of a newly created one
Atm only order 1 interactions
2.7.3. net_sim Module¶
The Net Simulator is divided into a Configuration, Net and a Result object.
After configuration of the network it can be instantiated by calling Net(conf) with a valid configuration conf. This creates eg. random connectivity so that the simulation with the same network can be repeated multiple times.
c = ni.model.net_sim.SimulationConfiguration()
c.Nneur = 10
net = ni.model.net_sim.Net(c)
print net
net.plot_firing_rates()
'ni.model.net_sim' Simulation Setup
Timerange: (250, 10250)
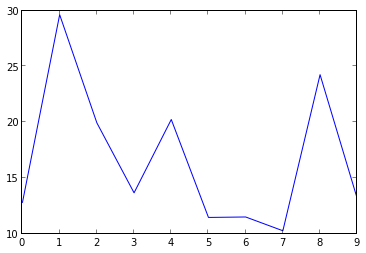
10 channels with firing rates:
[12.815928361, 29.6328550796, 19.9415819867, 13.6710936491, 20.242131795, 11.4661487294, 11.5071338947, 10.2727521514, 24.2587596858, 13.1497981307]
Firing Rates plot
for i in range(1,11):
print i
res1 = net.simulate()
res1.plot_firing_rates()
plot(numpy.array([r.num_spikes_per_channel for r in net.results]))
plot([0]*len(net.results))
- class ni.model.net_sim.Net(config)[source]¶
The Net Simulator class. Use with an Configuration instance.
- class ni.model.net_sim.SimulationConfiguration[source]¶
Configures the simulation. The default values are:
Nneur = 100 sparse_coeff = 0.1 Trial_Time = 1000 prior_epoch = 250 Ntrials = 10 Nsec = Ntrials*Trial_Time/1000 Ntime = Nsec*1000 eps =0.1 frate_mu = 1.0/25.0 Nhist = 50 output = False rate_function = False
2.7.4. create_design_matrix_vk Module¶
- ni.model.create_design_matrix_vk.computeCovariate(index, o, C, V1)[source]¶
Computes a row of the designMatrix corresponding to a certain covariate.
- ni.model.create_design_matrix_vk.create_design_matrix_vk(V1, o)[source]¶
Fills free rows in the current design matrix, deduced from size(mD) and len(freeCov), corresponding to a single covariate according to the spline bases of Volterra kernels. The current kernel(s) and the respective numbers of covariates that will be computed for each kernel is deduced from masterIndex by determining the position in hypothetical upper triangular part of hypercube with number of dimensions corresponding to current kernel order. Using only the ‘upper triangular part’ of the hypercube reflects the symmetry of the kernels which stems from the fact that only a single spline is used as basis function.
saves covariate information in cell array ‘covariates’, format is {kernelOrder relativePositionInKernel productTermsOfV1}
Anpassung für Gordon: masterIndex, log, C, mD, freeCov werden berechnet statt übergeben.
- ni.model.create_design_matrix_vk.detKernels(freeCov, masterIndex, oCov, mOrder, C)[source]¶
Determines from the number of free slots in Designmatrix len(freeCov) and the current masterIndex how many covariates for which Volterra coefficient can be computed. Updates model order mOrder.
- ni.model.create_design_matrix_vk.detModelOrder(masterIndex, C)[source]¶
Determines model order and corresponding number of covariates.
2.7.5. create_splines Module¶
- ni.model.create_splines.N(u, i, p, knots)[source]¶
Compute Spline Basis
Evaluates the spline basis of order p defined by knots at knot i and point u.
- ni.model.create_splines.augknt(knots, order)[source]¶
Augment knot sequence such that some boundary conditions are met.
- ni.model.create_splines.create_splines(length, nr_knots, remove_last_spline, fn_knots)[source]¶
Generates B-spline basis functions based on the length and number of knots of the ongoing iteration. fn_knots is a function that computes the knots.
- ni.model.create_splines.create_splines_linspace(length, nr_knots, remove_last_spline)[source]¶
Generates B-spline basis functions based on the length and number of knots of the ongoing iteration
- ni.model.create_splines.create_splines_logspace(length, nr_knots, remove_last_spline)[source]¶
Generates B-spline basis functions based on the length and number of knots of the ongoing iteration
- ni.model.create_splines.spcol(x, knots, spline_order)[source]¶
Computes the spline colocation matrix for knots in x.
The spline collocation matrix contains all m-p-1 bases defined by knots. Specifically it contains the ith basis in the ith column.
- Input:
- x: vector to evaluate the bases on knots: vector of knots spline_order: order of the spline
- Output:
- colmat: m x m-p matrix
- The colocation matrix has size m x m-p where m denotes the number of points the basis is evaluated on and p is the spline order. The colums contain the ith basis of knots evaluated on x.
2.7.6. backend_elasticnet Module¶
This module provides a backend to the .ip model. It wraps the sklearn.linear_model.ElasticNet / ElasticNetCV objects.